Housekeeping genes are genes that are persistently expressed and believed to have a role in essential cellular biological processes. They are frequently employed as benchmarks for normalization in molecular biology research. Prior studies mostly examined the stability of housekeeping genes in terms of tissue expression difference, rather than considering individual variability. In this study utilizing the GTEx datasets, our main focus is to explore consistently expressed housekeeping genes and utilize the Gini index to analyze patterns of expression variability not just within tissues but also across human individuals. The Gini index is an empirical measure utilized in economics to quantify the level of income inequality. The Gini coefficient / Gini index quantifies the disparity between the values of a frequency distribution, such as income levels. A Gini coefficient of 0 indicates absolute equality, in which the values of all wealth or income are identical. Conversely, a Gini coefficient of 1 signifies the greatest possible inequality among values, wherein a solitary individual possesses the entire income and none of the others do.
We computed the Gini index-subject using the entire 16,000 donor GTEx data for protein-coding genes. Additionally, we calculated the Gini index-tissue to assess the variability in gene expression among donor subjects within each tissue type. Ultimately, we selected 27 significant organs for the purpose of studying protein-coding genes involved in housekeeping functions. We combined the Gini index-subject and Gini index-tissue-based housekeeping genes to create alternate lists of housekeeping protein-coding genes. This knowledge on housekeeping genes may be advantageous for future study of integrated bioinformatic biomarker data.

 529 Housekeeping Reference Genes (Gini index-tissue < 0.2 in all 27 tissue subtypes)
529 Housekeeping Reference Genes (Gini index-tissue < 0.2 in all 27 tissue subtypes)
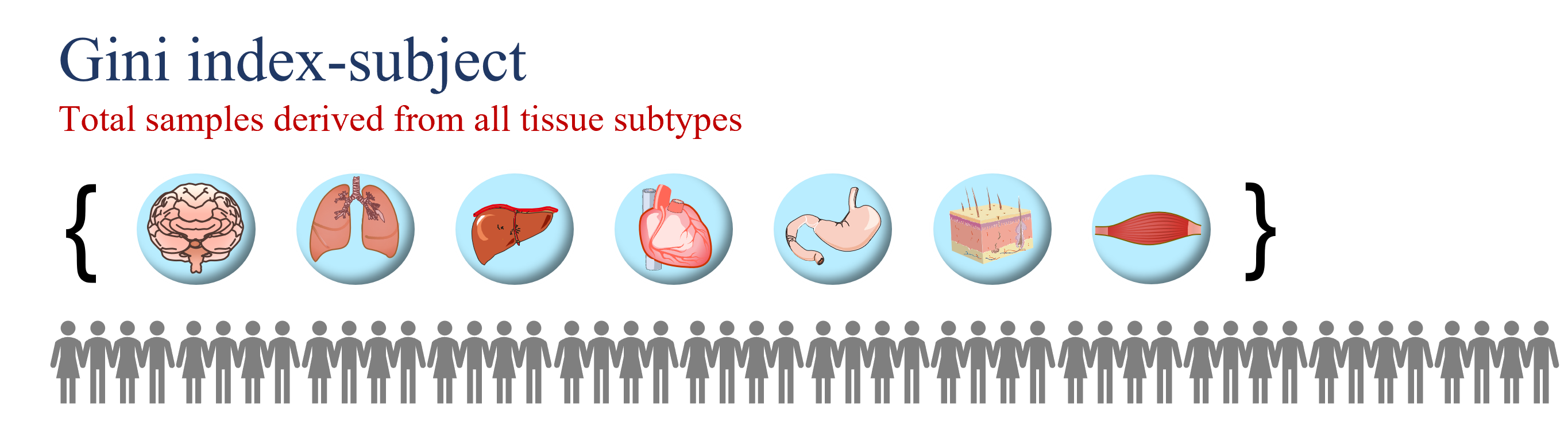
 335 Housekeeping Reference Genes (Gini index-subject < 0.2 in combined human subjects)
335 Housekeeping Reference Genes (Gini index-subject < 0.2 in combined human subjects)
 18,439 Genes (listed by chromosome locations):
18,439 Genes (listed by chromosome locations):